The Metabolomics & Systems Toxicology Laboratory is highly active in a series of coordinated research activities, from innovating new metabolomics technologies and methods to enable more informative toxicology studies, to testing those methods in research projects, through to translating the approaches as novel solutions for our industry and regulatory collaborators working in chemical safety science. Broad research areas are introduced below, and individual projects are described in Team Members & Projects.
Systems Toxicology
By exploiting the exceptional capability of metabolomics to conduct untargeted measurements of small molecule biochemistry, our Metabolomics & Systems Toxicology Laboratory specialises in discovering toxicity pathways of chemicals, drugs and nanomaterials in human and environmental toxicology. This strategy is applied to discover and validate metabolic Key Events to accelerate the creation of Adverse Outcome Pathways (AOPs). Related research projects include the applications of metabolomics for chemical grouping and read-across, and to probe the metabolism of the exposure chemicals, which we term untargeted toxicokinetics.
Several toxicology projects are undertaken in collaboration with Professor John Colbourne and his group (j.k.colbourne@bham.ac.uk)

Metabolomics method development
Underpinning this translational toxicology research, our Metabolomics & Systems Toxicology Laboratory also focuses on developing analytical and bio-/chemo-informatic methods in metabolomics. This includes novel methods to automate sample preparation using liquid handling robots through to developments in high resolution mass spectrometry, all focused towards applications in higher throughput, higher sensitivity toxicology.
Informatics research projects include optimising standardisable methods to process mass spectrometry data and to identify metabolites, and developing transparent and reproducible Galaxy workflows. Several informatics and computational projects are undertaken in collaboration with Dr Ralf Weber and his group (r.j.weber@bham.ac.uk).

Deep Metabolome Annotation
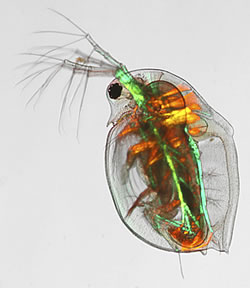
Deep Metabolome Annotation (DMA) is a flagship project in the Metabolomics & Systems Toxicology Laboratory, which is attempting the most extensive experimental characterisation of a complex model organism metabolome to date, utilising a range of analytical methods from LC-MS, GC-MS, DIMSn to NMR spectroscopy. Daphnia magna is the first organismal metabolome to be deeply annotated, which is a freshwater crustacean that is a sentinel species for ecotoxicology and more recently adopted as an NIH model organism for human health.
Metabolomics Standardisation in Regulatory Toxicology
To facilitate the translation of metabolomics into regulatory toxicology, projects are underway with the European Centre for Ecotoxicology and Toxicology of Chemicals (ECETOC) and the OECD to define best practice for analytical and computational metabolomics, including method performance criteria and minimal reporting standards.
The first guidance document has been published for the Metabolomics standaRds Initiative in Toxicology (MERIT) project, and can be found in our Nature Communications article here.
An OECD Guidance Document describing a metabolomics reporting framework (MRF) is nearing completion, as described here.
