The Drosophila Neurotrophin system is formed of:
- Drosophila Neurotrophin (DNT) ligands encoded by the spätzle paralogue gene family. Neurotrophin function has been demonstrated for DNT-1 (also known as Spz-2), DNT-2 (also known as Spz-5) and Spz-1.
- Toll receptors, which are expressed in the CNS and function as Neurotrophin receptors, maintaining neuronal survival during connectivity.
- Kinase-less Trk-Like receptors, encoded by the kekkon (kek) gene family. Keks share with Trk receptors the extracellular, ligand-binding domain but they lack the typical Tyrosine-kinase domain of most mammalian Trks.
The ligands are evolutionarily conserved. The receptors are distinct in Drosophila vs. mammals, or perhaps equivalent receptors may also be involved in mammals. Ultimately, the outcome is analogous: the regulation of cell survival, cell death and synaptic plasticity. To find out more, please read below and the following two pages.
Drosophila Neurotrophins
The mammalian neurotrophin (NT) protein family – formed of NGF, BDNF, NT3 and NT4 – underlies most aspects of nervous system development and function in the mammalian and human brain. They link structure and function in the brain, conferring it with the plasticity that enables it to make adjustments during growth and during learning. NTs regulate neuronal survival, cell proliferation, neuronal differentiation, axonal patterns, dendritic aroborisation, connectivity, synaptogenesis, synaptic transmission, learning and memory. Many neurodegenerative and psychiatric diseases result form alterations in neurotrophin function.
We have shown that a NT family in Drosophila formed by DNT1, DNT2 and Spz functions in nervous system development to adjust neuronal number during connectivity in the fly (Zhu et al 2008 PLoS Biol). Thus, a common mechanism is shared across the animals for nervous system formation.
DNT1 was identified as a BDNF homologue using bioinformatics tools developed to detect structural conservation despite sequence divergence (Zhu et al 2008 PLoS Biol). DNT1 is a paralogue of spz, and a further paralogue, DNT2, was identified as closest to DNT1. Spz has been shown by others to be structurally related to the mammalian neurotrophin family, as having an NGF domain (De Lotto & De Lotto 1998, Mizuguhi et al 1998), Spz crystals can be aligned with those of NGF (Arnot et al 2010) and binding of Spz to Toll resembles that of NGF to p75NTR. DNTs have the cystine-knot domain charactersitic of mature NTs, distinct from those of other growth factors like TGFb. The 6 Cysteines and their characteristic spacing, as well as other key residues, are conserved in NTs homologues throughout the invertebrates as well as vertebrates and mammals.
Like mammalian NTs, DNTs can also be secreted as pro-DNTs or cleaved mature DNTs. Cleavage of DNT1 and 2 is by furins, at conserved furin sites as in mammalian NTs (Foldi, Anthoney et al 2017, JCB).
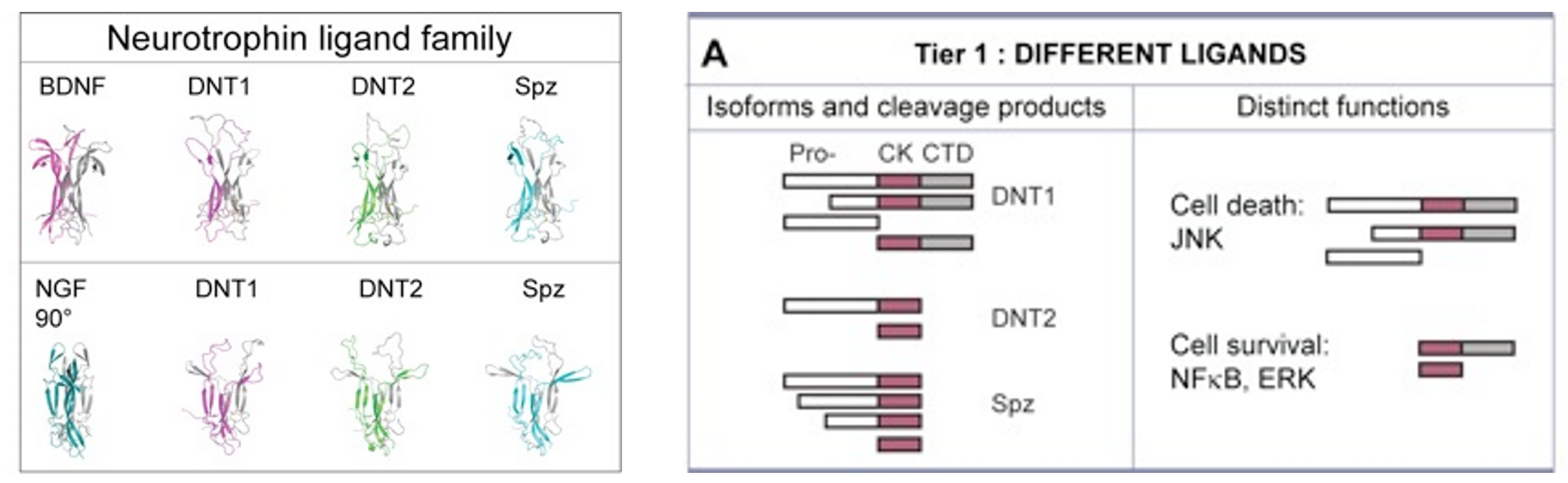
There is also functional conservation. Loss of DNT function leads to neuronal apoptosis in the Drosophila embryonic CNS, and over-expression of mature DNTs rescues naturally occurring cell death. Loss and gain of DNT1 function also alter targeting by motoraxons to the muscles (Zhu et al 2008). DNT1, DNT2 and Spz are all involved in embryonic CNS development, where they have partially redundant functions (mostly DNT1 and DNT2), and complementary functions (Spz vs. DNT1) (Zhu et al 2008). The expression of spz complements those of DNT1 and DNT2 in the embryonic muscles, and loss or gain of DNT1 and DNT2 function affect targeting by a set of ventral motoneurons distinct from those affected by alterations in Spz function. DNT1, DNT2 and Spz are required for synaptogenesis (Sutcliffe et al 2013). Pro-DNT1 can promote induce JNK signalling and promote cell death. Cleaved DNT1 and 2 can induce ERK and NFkB signalling and promote neuronal survival (Foldi, Anthoney 2017 JCB).
We collaborate with structural biologists and biochemists in the team of Prof. Nick Gay (Department of Biochemistry, University of Cambridge) to solve the mechanistic questions.
We look at all stages, from embryo to adult, and our approaches include classical genetics, molecular biology, cell culture, imaging and behaviour.
See review of our work by Keeler and Deppmann (2017) J Cell Biol.


