The ACM group has six core objectives focused on metabolism research, analytical chemistry and metabolomics.
1. Development and validation of innovative untargeted and semi-targeted UHPLC-MS assays which provide broad coverage of human metabolomes and which allow reproducible data collection for small and large studies
The group has a focus on the development of UHPLC-MS assays to allow the ‘global’ detection of metabolites in mammalian biofluids, tissues and cells in both small (<100 samples), medium (100-1000 samples) and large (>1000 samples) studies. In close association with Phenome Centre Birmingham, three assays have been developed to maximise metabolome coverage while providing high-quality data in medium-throughput assays (15-minute analysis times). Optimisation of sample preparation for different sample types has also been performed.
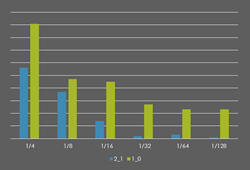
The majority of current UHPLC-MS assays apply a 2.1mm i.d. analytical column because sample volume/mass is not limited. To allow the analysis of smaller volume biofluids (e.g. tears) and lower mass tissues (e.g. needle biopsies), the group are developing higher sensitivity UHPLC-MS assays using 1.0mm i.d. analytical columns. This research is in collaboration with the Defence Science and Technology Laboratories (DSTL) in the UK.
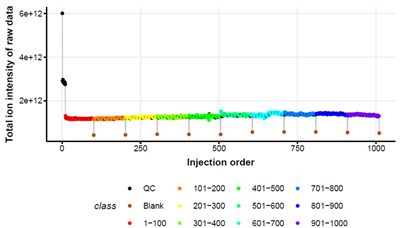
The collection of high-quality data is hugely important and our goals include the development of assays which allow data collection for 1000s of samples. We apply pooled QC samples and bioinformatics workflows to assess data filter data and report the quality of data.
Professor Dunn is chairman of the coordinating committee of the metabolomics Quality Assurance and quality Control Consortium (mQACC).
2. Development and validation of targeted UHPLC-MS assays for application in metabolism studies
Untargeted and semi-targeted metabolomics assays are applied as a discovery tool when the metabolites of biological or clinical interest are not known. However, these are research tools which will never be applied in a clinical translational environment and instead targeted assays measuring small numbers of metabolites would be the route for translation from bench to bedside. Targeted assays are developed and validated according to FDA recommendations in Phenome Centre Birmingham. Examples of the types of assays which are developed and validated are one carbon metabolism, caffeine and its metabolites, gut permeability tests applying sugars and sphingosine-1-phosphate.
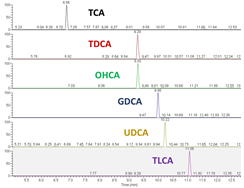
3. Development and validation of innovative analytical chemistry assays and computational resources for the annotation or identification of metabolites in untargeted and semi-targeted assays
Untargeted and semi-targeted metabolomics assays collect data without knowing the chemical identity of all or some of the metabolites detected. Instead data is collected to allow unidentified metabolites to be annotated or identified after data collection. Without metabolite annotation and identification, no biological knowledge can be derived and this process has been defined as a bottleneck. Our research has:
- characterised the complexity of data collected, for example, the chemical complexity of electrospray ionisation and the data generated
- created software tools for metabolite annotation based on full-scan MS1 data
- characterises and optimised the collection of MS/MS data to maximise the number of metabolites with high quality MS/MS or MSn datacharacterised the metabolite composition of human and mammalian biofluids and tissues to generate ‘Detectable Annotation and Identification of Metabolome’ (DAIM) computational resources
Some of this research is in collaboration with Professor Mark Viant, Dr Ralf Weber and Thermo Scientific.
Professor Dunn co-chairs the Metabolite Identification Task group of the Metabolomics Society.

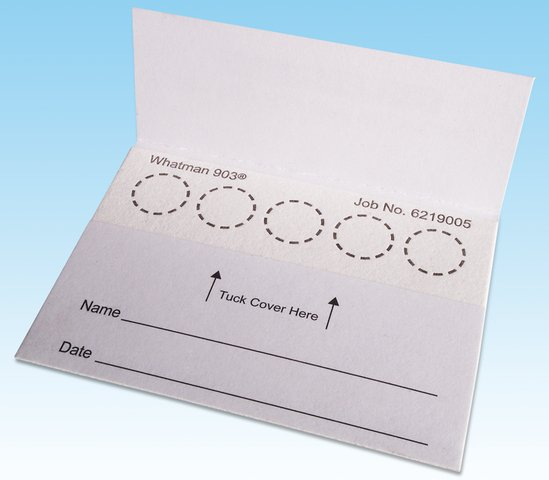
4. New approaches to sample collection away from the clinic including dried blood spots
The collection of biofluids from humans normally occurs in a clinical environment such as a doctor’s surgery or a hospital because collection is invasive and/or rapid sample processing is required. The group are investigating and applying new approaches to collect samples away from a clinical environment and by the subjects being studied. This includes the collection of dried blood spots which have been shown to be stable at room temperature for up to one month. Innovative sample analysis assays are also required and these include Liquid Extraction Surface Analysis (LESA). This research is in collaboration with Thermo Scientific and Professor Helen Cooper.
5. The study of metabolism in humans across the life course and related to human ageing and disease
Metabolites have many important roles in how human diseases develop and progress including in metabolic changes, in signalling and regulation and in cell membranes. The group applies metabolomics research tools to study biofluids, tissues and cells to (1) mechanistically understand how diseases develop and progress and (2) to develop biomarker panels to be applied to stratify patients based on risk of developing a disease, risk of disease progression and response to treatments.
Examples of the clinical areas we collaborate in are:
- Cancer including MGUS/multiple myeloma, CLL and childhood brain tumours with Professor Chris Bunce, Dr Helen Parry and Professor Andrew Peet
- Endocrinology, including steroid hormone dysfunction and Polycystic Ovarian Syndrome (PCOS) with Professor Wiebke Arlt and acromegaly with Dr Niki Karavanti.
- Hypertriglyceriaemia and APOC3 with Professor Nic Timpson
- Burn trauma with Professor Janet Lord
- Childhood cardiac disease with Dr Nigel Drury
- Liver transplantation with Dr Simon Afford

6. The study of metabolism in healthy humans during dietary or exercise interventions and in relation to circadian rhythms and their dysregulation
Metabolites also play important roles in how we respond to environmental and lifestyle factors including diet, exercise and circadian rhythms. The group applies metabolomic research tools to study metabolic responses to exercise (with Dr Sarah Aldred and Dr Gareth Wallis), diet (with Professor Georg Lietz) and circadian rhythms (with Prof. Debra Skene and Dr Robert Dallmann).

