The BEAMS (Birmingham mEtabolite Annotation for Mass Spectrometry) package includes several automated and seamless computational modules that are applied to putatively annotate metabolites detected in untargeted ultra (high) performance liquid chromatography-mass spectrometry or untargeted direct infusion mass spectrometry metabolomic assays in a single and automated process. All reported metabolites are annotated to level 2 or 3 of the Metabolomics Standards Initiative (MSI) reporting standards (as defined by Sumner et al. Proposed minimum reporting standards for chemical analysis, Chemical Analysis Working Group (CAWG), Metabolomics Standards Initiative (MSI). Metabolomics. 2007, 3(3): 211–221).
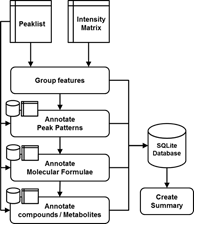
The package is highly flexible to suit the diversity of sample types studied and mass spectrometers applied in untargeted metabolomics studies. The user can use the standard reference files included in the package or can develop their own reference files.
The package is available to be used by computational experts (in python and Galaxy) and by laboratory researchers who are not experts in bioinformatics (using a GUI interface)

